Tutorial
FTFlex maps proteins while considering side chain flexibility. This is a multi-stage process that requires user input twice. The work flow for the server is as follows:
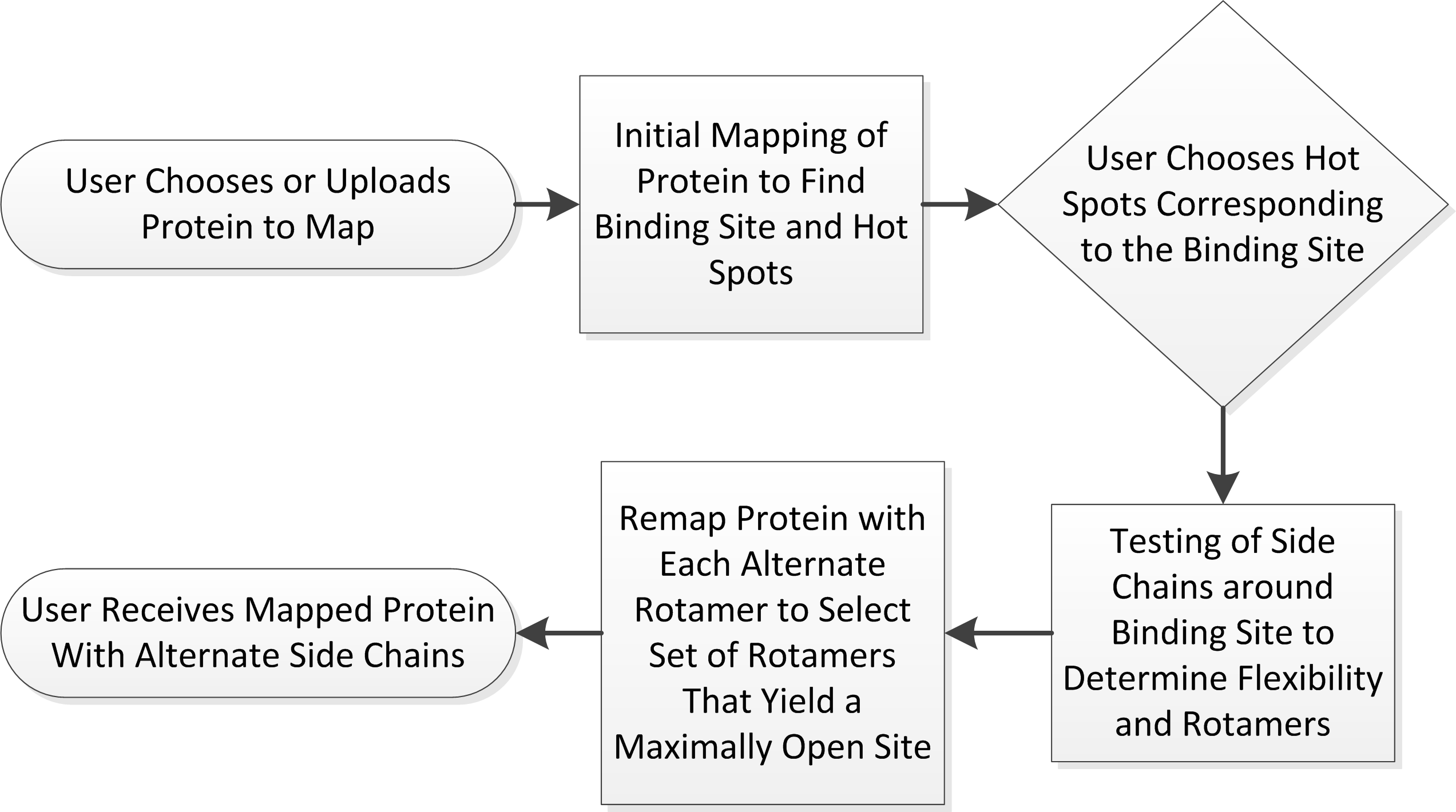
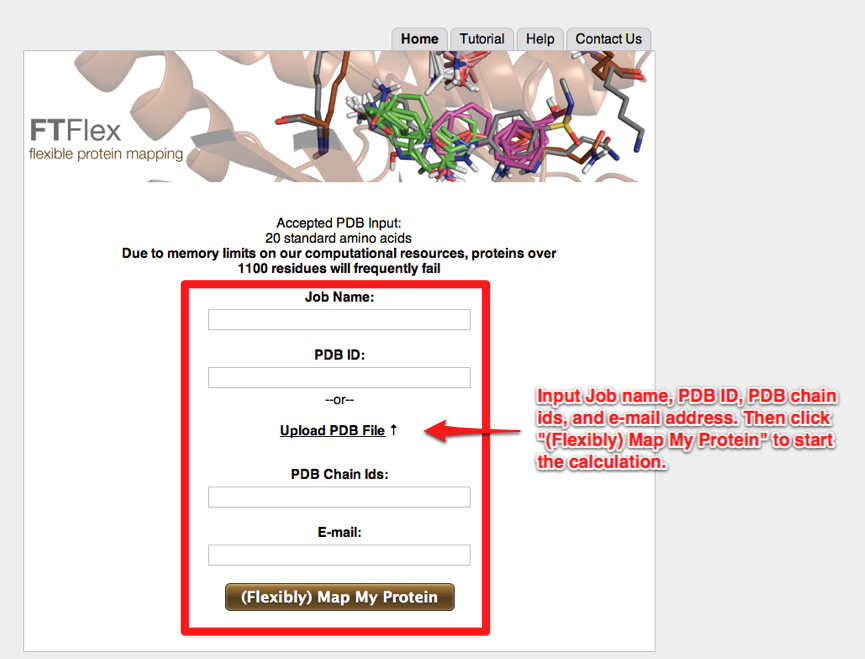
- A pdb file is submitted to the server by a user. On the “Home” tab, the user must input a job name, a PDB id, protein chain(s) to be mapped, and (optionally) an email address. The protein structure is then mapped using the same methods as the FTMap server. If no e-mail is provided, the user must keep track of the link provided post-submission to follow their job.
- If an e-mail address is provided, the initial mapping results will be returned to the user via e-mail along with a link that will allow the user to input information for the next stage of FTFlex. If no e-mail is provided, the user must keep track of the job using the provided link, which will be updated when the initial mapping finishes. On the provided page, the user must select the ligand binding region, or other region of interest, by specifying the consensus sites that define this region. The binding site will generally correspond to the region with the largest number of high-ranking probe clusters. After the user selects the applicable consensus sites, the server will search for flexible side chains within a 5 Å radius of those sites.
Note: The user can choose certain side chains to definitely not move at this stage. For example, if the user would like FTFlex to ignore Trp127 on chain B, the user would enter B-127.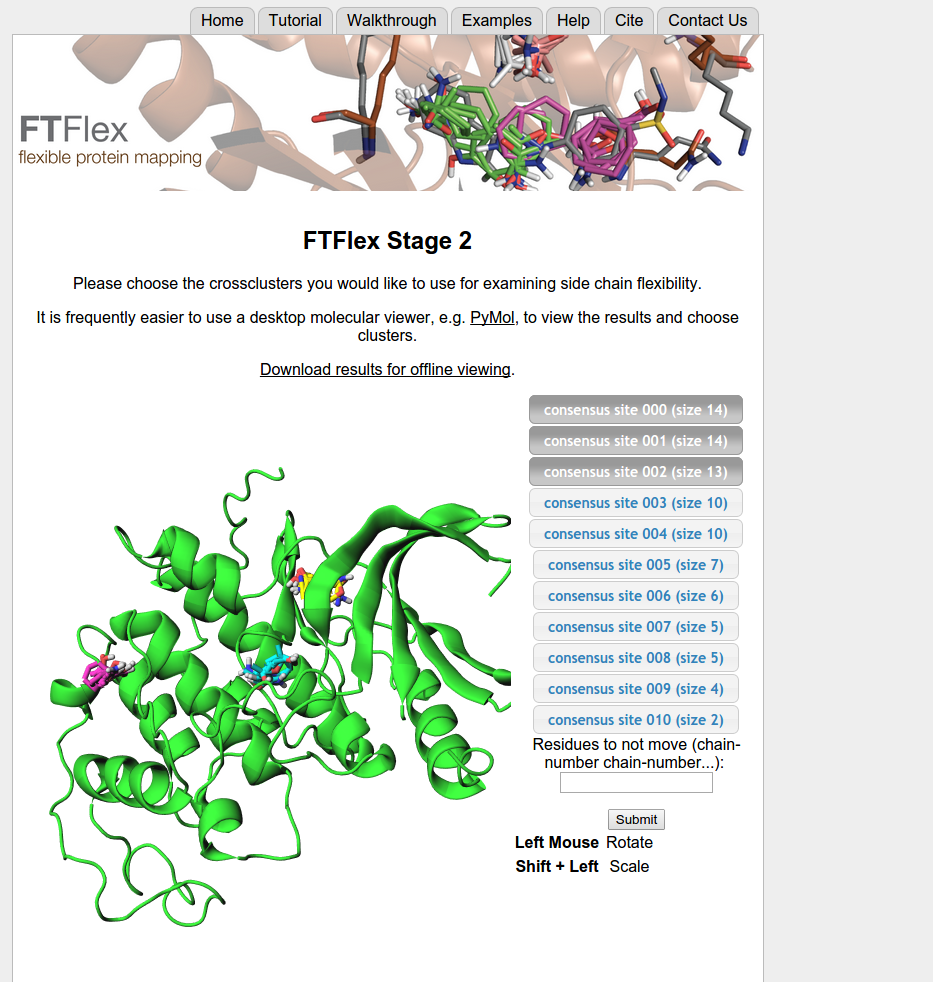
- The FTFlex server will individually generate and test rotamers for all side chains within the 5 Å radius of the selected probe clusters. The server will determine the set of rotamers for which the number of probe clusters within the defined binding site is maximized, thereby indicating a maximally opened binding site. The length of this step will depend on the number of possible rotamers, hence the time that this step takes is highly variable.
- A final email is sent to the user when the final conformation is determined. This email will contain the mapping results and information regarding which residues, if any, were moved to obtain the final mapping results. If no e-mail address was provided, the user may track their job online and download the results when the job is complete.